?mean()4 Basics of R
“Confusion is part of programming.”
– Felienne Hermans
4.1 Getting help
Probably the most basic thing to know is how to get help in R. Besides a quick Google search or asking ChatGPT, R also provides a help function. The help function can be accessed using ?.
The code above will open the Help pane, which explains what the function mean() does.
4.2 Executing the code
The codes can be typed either in R Script or Console. The code in R Scripts can be executed by placing the cursor at any line of the codes and clicking Ctrl + Enter in Windows and Cmd + Enter in Macs.
# Example 1: A single line of code
mean(1:10)
# Example 2: A multiple lines of code
c(1:10, 10.6, 11.9) |>
mean() |>
round(digits = 1)In the second example (example of multiple lines of code), a cursor can be placed at any line of code. Additionally, the codes in the Console can be executed by clicking Enter only. If you want to run any code in this book, you should copy and paste it into the R Script instead of the Console, especially if there are multiple lines of code. Notably, the codes should be run in sequence unless the current line of codes is independent from the previous line of codes.
4.3 Data types in R
There are a few data types in R:
- Numeric
- Integer
- Logical
- Character
- Complex
Let us see the example in R:
For the numeric:
x1 <- 11
x2 <- 11.9
class(x1); class(x2)[1] "numeric"[1] "numeric"Both numbers are recognised as numeric in R. For integers, the number should be denoted by ‘L’ to be recognised as an integer.
x3 <- 11L
class(x3)[1] "integer"For logical values, the boolean operators such as ‘FALSE’ and ‘TRUE’ are examples of logical values.
x4 <- c(TRUE, FALSE)
class(x4)[1] "logical"Next, we have character values.
x5 <- c("fruit", "apple")
class(x5)[1] "character"Lastly, we have complex values. The type of data is usually used to store numbers and imaginary components (for example, i in the code below).
x6 <- 9 + 3i
class(x6)[1] "complex"It is important to note the data type for each value as the function for numeric values can only be applied to numeric values. For example, if we want to find a mean value.
numeric_val <- 1:10 #list out all numbers between 1 and 10
numeric_val [1] 1 2 3 4 5 6 7 8 9 10character_val <- letters[1:10] #list out the first 10 alphabets
character_val [1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j"Now, let us try applying the mean function to both data.
mean(numeric_val)[1] 5.5mean(character_val)Warning in mean.default(character_val): argument is not numeric or logical:
returning NA[1] NASo, R gives us a warning that the ‘character_val’ is not a numeric or logical value. Thus, the returning NA mean not available.
4.4 Data structure in R
There are a few data structures in R:
- Vector
- Matrix
- Array
- Data frame
- List
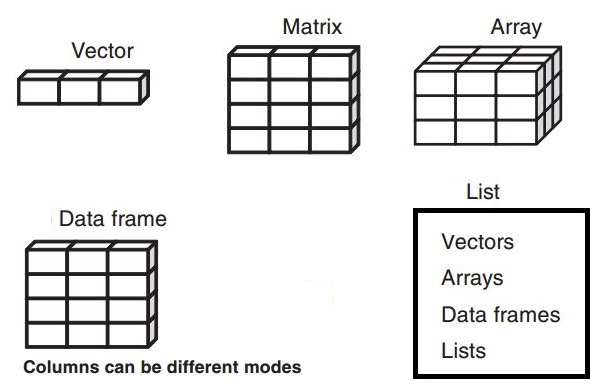
Depending on the fields, certain data structures are more common compared to others.
4.4.1 Vector
Vector is the most basic data structure in R. It can contain one data type at a time.
vec_data <- c(1, 2, 3, 4)
vec_data[1] 1 2 3 4The structure of the data can be checked using the function str().
str(vec_data) num [1:4] 1 2 3 4We can further confirm whether vec_data is a vector or not by using the is.vector() function.
is.vector(vec_data)[1] TRUEA TRUE result indicates that the data is a vector type.
4.4.2 Matrix
A matrix contains at least a single row and a single column. Contrary, to a vector which contains only a single row or a single column.
mat_data <- matrix(data = c(1, 2, 3, 4, 5, 6), nrow = 3, ncol = 2)
mat_data [,1] [,2]
[1,] 1 4
[2,] 2 5
[3,] 3 6By using str() function, we can see that the values are numerical and we have a matrix with 3 rows and 2 columns.
str(mat_data) num [1:3, 1:2] 1 2 3 4 5 6Next, we can confirm that our data is a matrix by using is.matrix().
is.matrix(mat_data)[1] TRUE4.4.3 Array
An array is quite similar to a matrix except that it can contain several layers of rows and columns.
arr_data <- array(data = c(1:6, 10:16), dim = c(2, 3, 2))
arr_data, , 1
[,1] [,2] [,3]
[1,] 1 3 5
[2,] 2 4 6
, , 2
[,1] [,2] [,3]
[1,] 10 12 14
[2,] 11 13 15As we can see, we have an array of 2 layers with each layer having 2 rows and 3 columns. By using the str() function, we can see that R recognises the array has integer values with 3 dimensions. The first dimension, 1:2 refers to the rows, the second dimension, 1:3 refers to the columns, and the last dimension, 1:2 refers to the layers.
str(arr_data) int [1:2, 1:3, 1:2] 1 2 3 4 5 6 10 11 12 13 ...is.array() can be used to ensure the data structure.
is.array(arr_data)[1] TRUE4.4.4 Data frame
A data frame is the extension of the matrix data structure. The difference between the former and the latter, the former contains the column names and each column may contain different data types.
df_data <- data.frame(
ID = 1:5,
Name = c("Mamat", "Abu", "Ali", "Chong", "Eva"),
Age = c(25, 30, 22, 35, 28),
Score = c(89, 95, 76, 88, 92)
)
df_data ID Name Age Score
1 1 Mamat 25 89
2 2 Abu 30 95
3 3 Ali 22 76
4 4 Chong 35 88
5 5 Eva 28 92We can further check the data structure and type using str(). So, our data structure is a data frame, consisting of 4 columns; the first column is an integer, the second column is a character, third and fourth columns are numeric.
str(df_data)'data.frame': 5 obs. of 4 variables:
$ ID : int 1 2 3 4 5
$ Name : chr "Mamat" "Abu" "Ali" "Chong" ...
$ Age : num 25 30 22 35 28
$ Score: num 89 95 76 88 92We can double-check the data structure using is.data.frame().
is.data.frame(df_data)[1] TRUE4.4.5 List
Lastly, we have a list. So, the list is a more advanced data structure in which we can have different data structures in a data structure. Let us see the example of a list in which we combine the previous vector and matrix data structures.
list_data <- list(
"vector" = vec_data,
"matrix" = mat_data
)
list_data$vector
[1] 1 2 3 4
$matrix
[,1] [,2]
[1,] 1 4
[2,] 2 5
[3,] 3 6We can further assess each data in the list using the $ symbol.
list_data$vector[1] 1 2 3 4By using str(), we see our data is a list with 2 elements or components; a vector and matrix.
str(list_data)List of 2
$ vector: num [1:4] 1 2 3 4
$ matrix: num [1:3, 1:2] 1 2 3 4 5 6To further confirm our data is a list, we can use is.list().
is.list(list_data)[1] TRUE4.5 Data in R
R itself contains the internal data that you can load for practice or other purposes. You can run data() in the Console to see what are the data available in R.
data()So, for example, if you want to load any of these data, you can type the name of the datasets. You will see under the Environment pane, the chickWeight dataset is loaded to your environment.
data("ChickWeight")Additionally, it is quite common that R packages have their own datasets as well. We will see this many times in this book going forward. Lastly, R is capable of reading different data formats.
| Format | R codes |
|---|---|
| .csv | read.csv() |
| .sav (SPSS) | haven::read_sav() |
| .xlsx (Excel) | readxl::read_excel() |
| .txt | read.table() |
| .dta (STATA) | haven::read_dta() |
The data formats that can be read are limited to the one listed in the table. Almost all data formats can be read in R. An efficient way to know whether R is capable of reading certain data formats by just a quick Google search.
For the rest of the chapter we going to the iris dataset, which is already available in R. To know more about this dataset, you can type the below code in the console.
?iris4.6 Packages
A package is a collection of functions and sample data that can be utilised for certain tasks. Certain functions in R are already loaded when you open R or RStudio. However, to use more advanced functions we need to install and load a package.
The packages can installed using install.packages(). For example, the code below will install the dplyr package which is commonly utilised for data wrangling and manipulation.
install.packages("dplyr")To use the installed packages, we need to load the packages using the library() function.
library(dplyr)Now, that we know what is a package, we might wonder where exactly these packages coming from.
The official packages in R are located in the Comprehensive R Archive Network (CRAN). This link contains all available packages in CRAN. Additionally, there is CRAN Task Views. At the time of this writing, CRAN contains 21,606 R packages intended for various tasks.
Furthermore, for those interested in bioinformatics, the related packages are located in the Bioconductor. At the time of this writing, Bioconductor contains 2,289 R packages related to bioinformatics.
In addition to CRAN and Bioconductor, there are unofficial R packages, which are usually located in GitHub and GitLab. There are probably thousands of these unofficial packages. For example, dmetar which is located in GitHub, contains R functions and codes to facilitate the conduction of meta-analyses.
More often than not, the official packages in CRAN also have their GitHub repositories in which they contain the latest development of R functions and codes before they going to be released in CRAN. So, current bugs and errors in the package are corrected first using the unofficial packages from GitHub or GitLab before they are eventually released to the CRAN repositories.
Several packages can help in installing the unofficial packages. The two most commonly used packages are devtools and remote (or at least I commonly used them).
First, we need to install the packages.
install.packages("devtools")
install.packages("remote")Next, we can install the unofficial packages.
devtools::install_github("MathiasHarrer/dmetar")
remotes::install_github("MathiasHarrer/dmetar")MathiasHarrer refers to the GitHub account or usually the author’s account on GitHub, followed by the name of the package. Depending on one’s preference, we can choose to use either devtools or remote.
4.7 Functions
A function in R is a block of code designed to perform a specific task. It consists of an argument, in which we need to supply it. For example, the mean() function is designed to find the mean or average across the numeric values. The numeric values are the argument that we need to supply to the function.
num_values <- c(5, 6, 8, 10)
mean(num_values)[1] 7.25The base R itself has numerous functions that are accessible to us. These base R functions can be used immediately once we open R (or RStudio or any other IDEs). In contrast, we also have R functions from the packages that we installed. First, we need to install the packages.
install.packages("tidyverse")We will learn about tidyverse in a little bit. Coming on to our current installation, once you install the package there are two ways to use the function inside the package. First, by loading the package, subsequently, all the functions in the package are ready to be used by us. For example below, bind_rows() is one of the functions from the dplyr package.
# Load the package
library(dplyr)
# Example of functions from dplyr package
bind_rows()
bind_cols()Secondly, we can use :: to access a single function that we are interested in. However, in this approach, only a single function is loaded.
# Example of dplyr functions
dplyr::bind_rows()
dplyr::bind_cols()As we can see, every time we want to call a function from the dplyr package we type dplyr:: as we do not load the package first.
4.8 Tidyverse package
We have learned that R packages contain a collection of functions and R codes that we utilise once we load the packages. So, tidyverse is an opinionated collection of R packages designed for data science (Wickham et al. 2019).
# List of all packages in tidyverse
tidyverse::tidyverse_packages(include_self = FALSE) |>
data.frame(Packages = _) Packages
1 broom
2 conflicted
3 cli
4 dbplyr
5 dplyr
6 dtplyr
7 forcats
8 ggplot2
9 googledrive
10 googlesheets4
11 haven
12 hms
13 httr
14 jsonlite
15 lubridate
16 magrittr
17 modelr
18 pillar
19 purrr
20 ragg
21 readr
22 readxl
23 reprex
24 rlang
25 rstudioapi
26 rvest
27 stringr
28 tibble
29 tidyr
30 xml2Table 4.2 below summarises common packages in tidyverse and its uses.
| Packages | Summary |
|---|---|
| ggplot2 | For data visualisation. |
| dplyr | Provides tools for data manipulation, including functions for filtering, selecting, grouping, and summarizing data. |
| tidyr | Specializes in data tidying, allowing users to transform datasets into a tidy format ready for analysis or visualisation. |
| readr | Used for reading rectangular data (e.g., CSV, TSV files) into R quickly and efficiently. |
| purr | Introduces a functional programming paradigm in R with tools for applying functions to data structures like lists and vectors. |
| tibble | Enhances data frames in R by making them more user-friendly with better printing options and stricter type checking |
| stringr | Facilitates consistent handling of strings, offering functions for string manipulation, pattern matching, and transformations. |
| forcats | Designed to work with categorical data (factors). |
Tidyverse is commonly utilised for data analysis and throughout this book, we going to use tidyverse functions numerous times.
4.9 Pipe operators
There are two types of pipe in R:
|>: this pipe is from base R, first introduced in R version 4.1.0.%>%: this pipe is from themagrittrpackage, which presentedtidyverse.
To use |>, we do not actually need to load anything as it is already available in base R. However, to use %>%, you need to load tidyverse. Certain tidyverse associated packages such as dplyr, forcats, and magrittr also load the %>%.
The main function of these pipe operators (regardless of which one we use) is to make our R codes more readable and intuitive. So, let us compare the codes without and with the pipe.
mean_value <- mean(
subset(
data.frame(value = 1:10, group = rep(c("A", "B"), each = 5)),
group == "A"
)$value
)
mean_value[1] 3Now, compare the codes with the pipe.
mean_value <- data.frame(value = 1:10, group = rep(c("A", "B"), each = 5)) |>
subset(group == "A") |>
with(mean(value))
mean_value[1] 3Basically, what we do in both R codes are:
Create a data frame with ten rows and two columns.
data.frame(value = 1:10, group = rep(c("A", "B"), each = 5))value group 1 1 A 2 2 A 3 3 A 4 4 A 5 5 A 6 6 B 7 7 B 8 8 B 9 9 B 10 10 B
Filter out the group column to value A.
data.frame(value = 1:10, group = rep(c("A", "B"), each = 5)) |> subset(group == "A")value group 1 1 A 2 2 A 3 3 A 4 4 A 5 5 ACalculate the mean.
mean(c(1, 2, 3, 4, 5))[1] 3
Coming back to both R codes, we can intuitively see that the codes with the pipe are more readable and clear compared to the other one. The codes with pipe can be read line by line, while the codes without the pipe need to be read inside out. As you can imagine, once our codes are more complex, the less readable the codes will become.
So, which pipe operators to choose?
In most cases, the differences are not significant enough to impact your code. Therefore, you can choose the pipe operator that best suits your preference or coding style. Most users will find that both options perform similarly for general tasks, so selecting one often comes down to familiarity or ease of use.
|> is a built-in pipe operator which can be used immediately without the need to load any package. %>% can be used once the tidyverse package is loaded. The latter pipe has short cut built-in in RStudio. For Windows user, Ctrl + Shift + M and for Mac user, Cmd + Shift + M. Additionally, we can set up the shortcut for |> or further change the shortcut for %>%, though, this will not be covered in this book.
4.10 Chapter summary
In this chapter, we learn about:
- Data, its types and structures in R.
- R packages and functions.
- Pipe operators.
4.11 Revision
What this
?actually do in R?List all the data types that we learn in this chapter.
List all the data structures that we learn in this chapter.
What is the difference between installing the package and loading the package?
Which one to choose between
|>and%>%?List three packages in
tidyverseand summarise their functionalities.